Enhancing gene regulation analysis with deep learning and DNA shape integration.
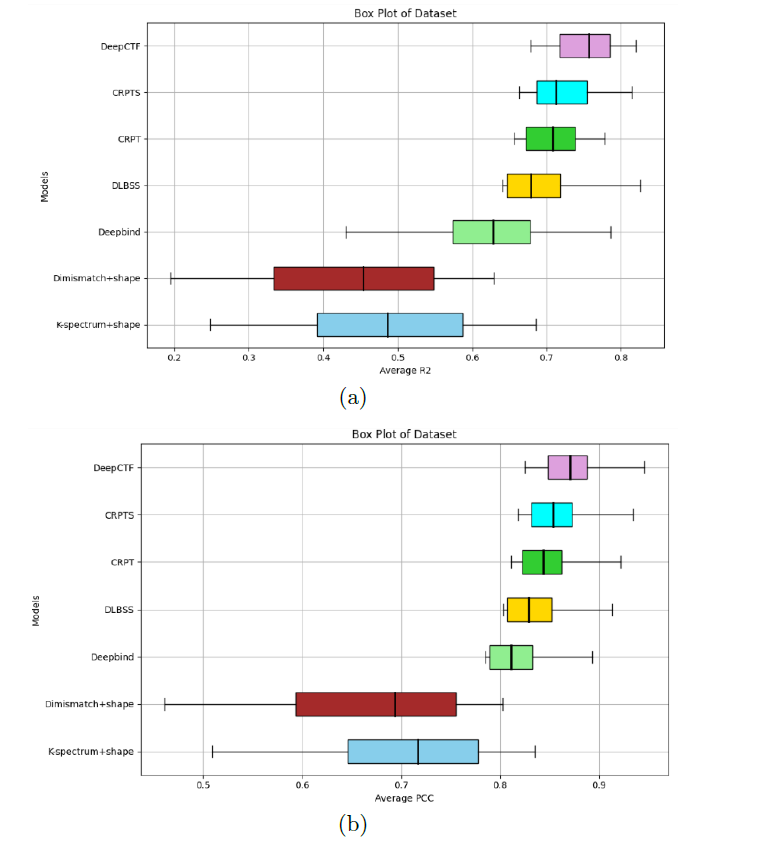
Problem Statement: Traditional TFBS prediction models struggle with feature extraction, long-range dependencies, and integrating DNA shape information. DeepCTF addresses these limitations by leveraging deep learning techniques.
Self-Attention Mechanism: Captures long-range dependencies for enhanced prediction accuracy.
Convolutional Neural Networks (CNNs): Identifies critical sequence motifs in DNA.
Bidirectional LSTM (BiLSTM): Captures sequential patterns and dependencies.
DNA Shape-Based Feature Augmentation: Incorporates DNA’s structural information.
Advanced Data Processing: Includes one-hot encoding, batch normalization, dropout, and L2 regularization.
Enhance TFBS predictions with DeepCTF's cutting-edge deep learning approach.
Get in Touch